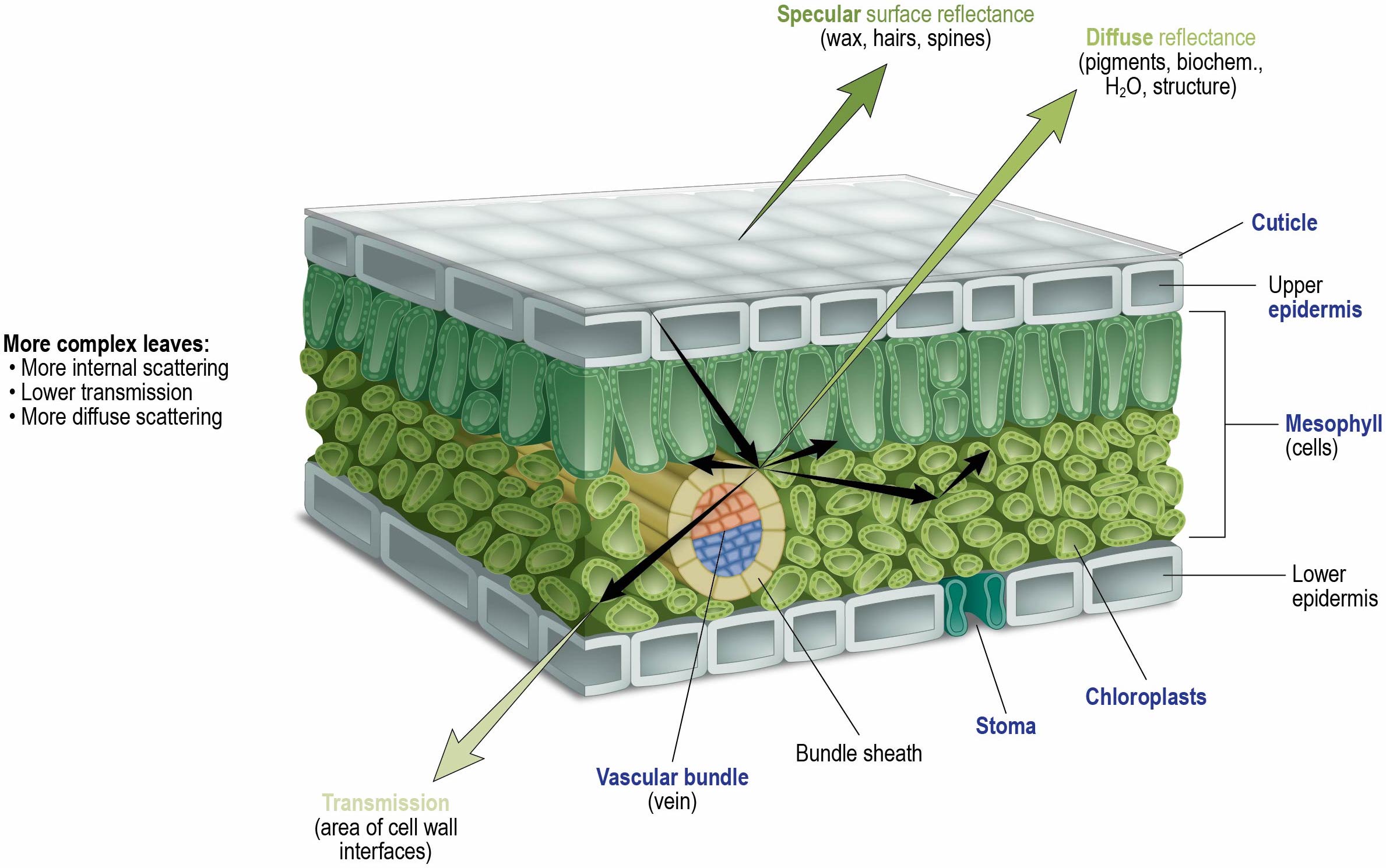
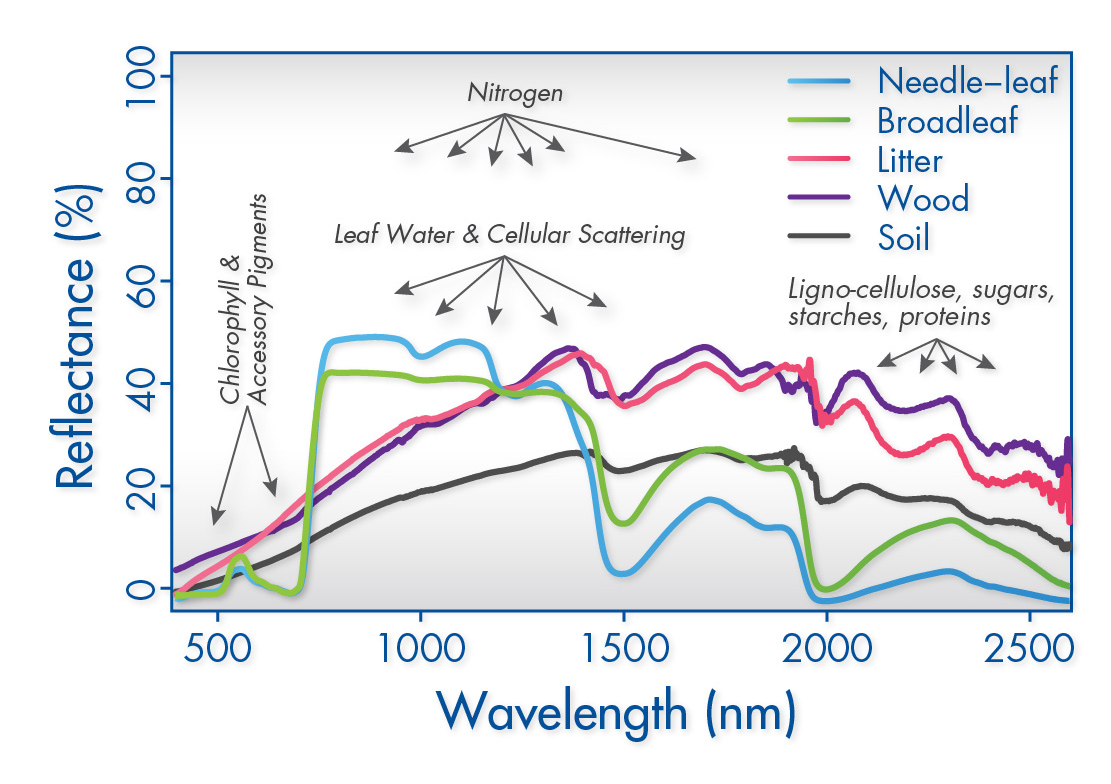
-
Notifications
You must be signed in to change notification settings - Fork 9
Example Tutorial #04: NEON CONUS dataset
Shawn P. Serbin edited this page Jun 19, 2024
·
1 revision
Spectra-trait PLSR example using leaf-level spectra and leaf mass per area (LMA) data from CONUS NEON sites
Shawn P. Serbin, Julien Lamour, & Jeremiah Anderson 2024-06-19
This is an R Markdown Notebook to illustrate how to retrieve a dataset from the EcoSIS spectral database, choose the “optimal” number of plsr components, and fit a plsr model for leaf-mass area (LMA)
list.of.packages <- c("pls","dplyr","here","plotrix","ggplot2","gridExtra","spectratrait")
invisible(lapply(list.of.packages, library, character.only = TRUE))## Warning: package 'pls' was built under R version 4.3.1
##
## Attaching package: 'pls'
## The following object is masked from 'package:stats':
##
## loadings
## Warning: package 'dplyr' was built under R version 4.3.1
##
## Attaching package: 'dplyr'
## The following objects are masked from 'package:stats':
##
## filter, lag
## The following objects are masked from 'package:base':
##
## intersect, setdiff, setequal, union
## here() starts at /Users/sserbin/Library/CloudStorage/OneDrive-NASA/Data/Github/spectratrait
## Warning: package 'plotrix' was built under R version 4.3.1
## Warning: package 'ggplot2' was built under R version 4.3.1
##
## Attaching package: 'gridExtra'
## The following object is masked from 'package:dplyr':
##
## combine
### Setup options
# Script options
pls::pls.options(plsralg = "oscorespls")
pls::pls.options("plsralg")## $plsralg
## [1] "oscorespls"
# Default par options
opar <- par(no.readonly = T)
# What is the target variable?
inVar <- "LMA_gDW_m2"
# What is the source dataset from EcoSIS?
ecosis_id <- "5617da17-c925-49fb-b395-45a51291bd2d"
# Specify output directory, output_dir
# Options:
# tempdir - use a OS-specified temporary directory
# user defined PATH - e.g. "~/scratch/PLSR"
output_dir <- "tempdir"## [1] "/private/var/folders/th/fpt_z3417gn8xgply92pvy6r0000gq/T/RtmpueELda"
URL: https://ecosis.org/package/fresh-leaf-spectra-to-estimate-lma-over-neon-domains-in-eastern-united-states
print(paste0("Output directory: ",getwd())) # check wd## [1] "Output directory: /Users/sserbin/Library/CloudStorage/OneDrive-NASA/Data/Github/spectratrait/vignettes"
### Get source dataset from EcoSIS
dat_raw <- spectratrait::get_ecosis_data(ecosis_id = ecosis_id)## [1] "**** Downloading Ecosis data ****"
## Downloading data...
## Rows: 6312 Columns: 2162
## ── Column specification ────────────────────────────────────────────────────────
## Delimiter: ","
## chr (10): Affiliation, Common Name, Domain, Functional_type, Latin Genus, ...
## dbl (2152): LMA, 350, 351, 352, 353, 354, 355, 356, 357, 358, 359, 360, 361,...
##
## ℹ Use `spec()` to retrieve the full column specification for this data.
## ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
## Download complete!
head(dat_raw)## # A tibble: 6 × 2,162
## Affiliation `Common Name` Domain Functional_type LMA `Latin Genus`
## <chr> <chr> <chr> <chr> <dbl> <chr>
## 1 University of Wiscon… black walnut D02 broadleaf 72.9 Juglans
## 2 University of Wiscon… black walnut D02 broadleaf 72.9 Juglans
## 3 University of Wiscon… black walnut D02 broadleaf 60.8 Juglans
## 4 University of Wiscon… black walnut D02 broadleaf 60.8 Juglans
## 5 University of Wiscon… black walnut D02 broadleaf 85.9 Juglans
## 6 University of Wiscon… black walnut D02 broadleaf 85.9 Juglans
## # ℹ 2,156 more variables: `Latin Species` <chr>, PI <chr>, Project <chr>,
## # Sample_ID <chr>, `USDA Symbol` <chr>, `350` <dbl>, `351` <dbl>,
## # `352` <dbl>, `353` <dbl>, `354` <dbl>, `355` <dbl>, `356` <dbl>,
## # `357` <dbl>, `358` <dbl>, `359` <dbl>, `360` <dbl>, `361` <dbl>,
## # `362` <dbl>, `363` <dbl>, `364` <dbl>, `365` <dbl>, `366` <dbl>,
## # `367` <dbl>, `368` <dbl>, `369` <dbl>, `370` <dbl>, `371` <dbl>,
## # `372` <dbl>, `373` <dbl>, `374` <dbl>, `375` <dbl>, `376` <dbl>, …
names(dat_raw)[1:40]## [1] "Affiliation" "Common Name" "Domain" "Functional_type"
## [5] "LMA" "Latin Genus" "Latin Species" "PI"
## [9] "Project" "Sample_ID" "USDA Symbol" "350"
## [13] "351" "352" "353" "354"
## [17] "355" "356" "357" "358"
## [21] "359" "360" "361" "362"
## [25] "363" "364" "365" "366"
## [29] "367" "368" "369" "370"
## [33] "371" "372" "373" "374"
## [37] "375" "376" "377" "378"
### Create plsr dataset
Start.wave <- 500
End.wave <- 2400
wv <- seq(Start.wave,End.wave,1)
Spectra <- as.matrix(dat_raw[,names(dat_raw) %in% wv])
colnames(Spectra) <- c(paste0("Wave_",wv))
sample_info <- dat_raw[,names(dat_raw) %notin% seq(350,2500,1)]
head(sample_info)## # A tibble: 6 × 11
## Affiliation `Common Name` Domain Functional_type LMA `Latin Genus`
## <chr> <chr> <chr> <chr> <dbl> <chr>
## 1 University of Wiscon… black walnut D02 broadleaf 72.9 Juglans
## 2 University of Wiscon… black walnut D02 broadleaf 72.9 Juglans
## 3 University of Wiscon… black walnut D02 broadleaf 60.8 Juglans
## 4 University of Wiscon… black walnut D02 broadleaf 60.8 Juglans
## 5 University of Wiscon… black walnut D02 broadleaf 85.9 Juglans
## 6 University of Wiscon… black walnut D02 broadleaf 85.9 Juglans
## # ℹ 5 more variables: `Latin Species` <chr>, PI <chr>, Project <chr>,
## # Sample_ID <chr>, `USDA Symbol` <chr>
sample_info2 <- sample_info %>%
select(Domain,Functional_type,Sample_ID,USDA_Species_Code=`USDA Symbol`,LMA_gDW_m2=LMA)
head(sample_info2)## # A tibble: 6 × 5
## Domain Functional_type Sample_ID USDA_Species_Code LMA_gDW_m2
## <chr> <chr> <chr> <chr> <dbl>
## 1 D02 broadleaf P0001 JUNI 72.9
## 2 D02 broadleaf L0001 JUNI 72.9
## 3 D02 broadleaf P0002 JUNI 60.8
## 4 D02 broadleaf L0002 JUNI 60.8
## 5 D02 broadleaf P0003 JUNI 85.9
## 6 D02 broadleaf L0003 JUNI 85.9
plsr_data <- data.frame(sample_info2,Spectra)
rm(sample_info,sample_info2,Spectra)### Create cal/val datasets
## Make a stratified random sampling in the strata USDA_Species_Code and Domain
method <- "dplyr" #base/dplyr
# base R - a bit slow
# dplyr - much faster
split_data <- spectratrait::create_data_split(dataset=plsr_data,approach=method, split_seed=2356812,
prop=0.8, group_variables=c("USDA_Species_Code","Domain"))
names(split_data)## [1] "cal_data" "val_data"
cal.plsr.data <- split_data$cal_data
head(cal.plsr.data)[1:8]## Domain Functional_type Sample_ID USDA_Species_Code LMA_gDW_m2 Wave_500
## 1 D08 broadleaf L2644 ACBA 44.18 0.04170800
## 2 D08 broadleaf L2646 ACBA 41.71 0.05067800
## 3 D08 broadleaf L2645 ACBA 40.66 0.04701700
## 4 D08 broadleaf P2639 ACBA 44.18 0.04125300
## 5 D03 broadleaf P0614 ACFL 52.91 0.03895800
## 6 D03 broadleaf L0609 ACFL 81.67 0.04186169
## Wave_501 Wave_502
## 1 0.04208700 0.04283700
## 2 0.05087600 0.05153500
## 3 0.04718200 0.04766500
## 4 0.04150300 0.04247100
## 5 0.03915100 0.03956200
## 6 0.04217802 0.04258768
val.plsr.data <- split_data$val_data
head(val.plsr.data)[1:8]## Domain Functional_type Sample_ID USDA_Species_Code LMA_gDW_m2 Wave_500
## 3 D02 broadleaf P0002 JUNI 60.77 0.043758
## 12 D02 broadleaf L0006 JUNI 42.54 0.044338
## 13 D02 broadleaf P0007 QUVE 106.57 0.015643
## 19 D02 broadleaf P0010 PRSE 78.82 0.033019
## 21 D02 broadleaf P0011 PRSE 86.09 0.024819
## 28 D02 broadleaf L0014 PRSE 67.11 0.040095
## Wave_501 Wave_502
## 3 0.044171 0.044869
## 12 0.044748 0.045294
## 13 0.015579 0.015431
## 19 0.033102 0.033245
## 21 0.024826 0.025045
## 28 0.040397 0.040864
rm(split_data)
# Datasets:
print(paste("Cal observations: ",dim(cal.plsr.data)[1],sep=""))## [1] "Cal observations: 4922"
print(paste("Val observations: ",dim(val.plsr.data)[1],sep=""))## [1] "Val observations: 1390"
cal_hist_plot <- ggplot(data = cal.plsr.data,
aes(x = cal.plsr.data[,paste0(inVar)])) +
geom_histogram(fill=I("grey50"),col=I("black"),alpha=I(.7)) +
labs(title=paste0("Calibration Histogram for ",inVar), x = paste0(inVar),
y = "Count")
val_hist_plot <- ggplot(data = val.plsr.data,
aes(x = val.plsr.data[,paste0(inVar)])) +
geom_histogram(fill=I("grey50"),col=I("black"),alpha=I(.7)) +
labs(title=paste0("Validation Histogram for ",inVar), x = paste0(inVar),
y = "Count")
histograms <- grid.arrange(cal_hist_plot, val_hist_plot, ncol=2)## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.

ggsave(filename = file.path(outdir,paste0(inVar,"_Cal_Val_Histograms.png")),
plot = histograms, device="png", width = 30, height = 12, units = "cm",
dpi = 300)
# output cal/val data
write.csv(cal.plsr.data,file=file.path(outdir,paste0(inVar,'_Cal_PLSR_Dataset.csv')),
row.names=FALSE)
write.csv(val.plsr.data,file=file.path(outdir,paste0(inVar,'_Val_PLSR_Dataset.csv')),
row.names=FALSE)### Format PLSR data for model fitting
cal_spec <- as.matrix(cal.plsr.data[, which(names(cal.plsr.data) %in% paste0("Wave_",wv))])
cal.plsr.data <- data.frame(cal.plsr.data[, which(names(cal.plsr.data) %notin% paste0("Wave_",wv))],
Spectra=I(cal_spec))
head(cal.plsr.data)[1:5]## Domain Functional_type Sample_ID USDA_Species_Code LMA_gDW_m2
## 1 D08 broadleaf L2644 ACBA 44.18
## 2 D08 broadleaf L2646 ACBA 41.71
## 3 D08 broadleaf L2645 ACBA 40.66
## 4 D08 broadleaf P2639 ACBA 44.18
## 5 D03 broadleaf P0614 ACFL 52.91
## 6 D03 broadleaf L0609 ACFL 81.67
val_spec <- as.matrix(val.plsr.data[, which(names(val.plsr.data) %in% paste0("Wave_",wv))])
val.plsr.data <- data.frame(val.plsr.data[, which(names(val.plsr.data) %notin% paste0("Wave_",wv))],
Spectra=I(val_spec))
head(val.plsr.data)[1:5]## Domain Functional_type Sample_ID USDA_Species_Code LMA_gDW_m2
## 3 D02 broadleaf P0002 JUNI 60.77
## 12 D02 broadleaf L0006 JUNI 42.54
## 13 D02 broadleaf P0007 QUVE 106.57
## 19 D02 broadleaf P0010 PRSE 78.82
## 21 D02 broadleaf P0011 PRSE 86.09
## 28 D02 broadleaf L0014 PRSE 67.11
par(mfrow=c(1,2)) # B, L, T, R
spectratrait::f.plot.spec(Z=cal.plsr.data$Spectra,wv=wv,plot_label="Calibration")
spectratrait::f.plot.spec(Z=val.plsr.data$Spectra,wv=wv,plot_label="Validation")
dev.copy(png,file.path(outdir,paste0(inVar,'_Cal_Val_Spectra.png')),
height=2500,width=4900, res=340)## quartz_off_screen
## 3
dev.off();## quartz_off_screen
## 2
par(mfrow=c(1,1))### Use permutation to determine the optimal number of components
if(grepl("Windows", sessionInfo()$running)){
pls.options(parallel = NULL)
} else {
pls.options(parallel = parallel::detectCores()-1)
}
method <- "firstPlateau" #pls, firstPlateau, firstMin
random_seed <- 2356812
seg <- 250
maxComps <- 20
iterations <- 40
prop <- 0.70
if (method=="pls") {
nComps <- spectratrait::find_optimal_components(dataset=cal.plsr.data, targetVariable=inVar,
method=method,
maxComps=maxComps, seg=seg,
random_seed=random_seed)
print(paste0("*** Optimal number of components: ", nComps))
} else {
nComps <- spectratrait::find_optimal_components(dataset=cal.plsr.data, targetVariable=inVar,
method=method,
maxComps=maxComps, iterations=iterations,
seg=seg, prop=prop,
random_seed=random_seed)
}## [1] "*** Identifying optimal number of PLSR components ***"
## [1] "*** Running permutation test. Please hang tight, this can take awhile ***"
## [1] "Options:"
## [1] "Max Components: 20 Iterations: 40 Data Proportion (percent): 70"
## [1] "*** Providing PRESS and coefficient array output ***"
## No id variables; using all as measure variables
## [1] "*** Optimal number of components based on t.test: 12"

dev.copy(png,file.path(outdir,paste0(paste0(inVar,"_PLSR_Component_Selection.png"))),
height=2800, width=3400, res=340)## quartz_off_screen
## 3
dev.off();## quartz_off_screen
## 2
### Fit final model
segs <- 100
plsr.out <- plsr(as.formula(paste(inVar,"~","Spectra")),scale=FALSE,ncomp=nComps,
validation="CV",
segments=segs, segment.type="interleaved",trace=FALSE,
data=cal.plsr.data)
fit <- plsr.out$fitted.values[,1,nComps]
pls.options(parallel = NULL)
# External validation fit stats
par(mfrow=c(1,2)) # B, L, T, R
pls::RMSEP(plsr.out, newdata = val.plsr.data)## (Intercept) 1 comps 2 comps 3 comps 4 comps 5 comps
## 29.174 18.644 18.115 15.657 12.508 11.978
## 6 comps 7 comps 8 comps 9 comps 10 comps 11 comps
## 11.766 11.220 10.941 10.226 9.934 9.580
## 12 comps
## 9.453
plot(pls::RMSEP(plsr.out,estimate=c("test"),newdata = val.plsr.data),
main="MODEL RMSEP",
xlab="Number of Components",ylab="Model Validation RMSEP",lty=1,col="black",
cex=1.5,lwd=2)
box(lwd=2.2)
pls::R2(plsr.out, newdata = val.plsr.data)## (Intercept) 1 comps 2 comps 3 comps 4 comps 5 comps
## -0.002151 0.590718 0.613614 0.711350 0.815784 0.831077
## 6 comps 7 comps 8 comps 9 comps 10 comps 11 comps
## 0.836995 0.851779 0.859055 0.876874 0.883790 0.891931
## 12 comps
## 0.894776
plot(pls::R2(plsr.out,estimate=c("test"),newdata = val.plsr.data), main="MODEL R2",
xlab="Number of Components",ylab="Model Validation R2",lty=1,col="black",
cex=1.5,lwd=2)
box(lwd=2.2)
par(opar)#calibration
cal.plsr.output <- data.frame(cal.plsr.data[, which(names(cal.plsr.data) %notin% "Spectra")],
PLSR_Predicted=fit,
PLSR_CV_Predicted=as.vector(plsr.out$validation$pred[,,nComps]))
cal.plsr.output <- cal.plsr.output %>%
mutate(PLSR_CV_Residuals = PLSR_CV_Predicted-get(inVar))
head(cal.plsr.output)## Domain Functional_type Sample_ID USDA_Species_Code LMA_gDW_m2 PLSR_Predicted
## 1 D08 broadleaf L2644 ACBA 44.18 53.09004
## 2 D08 broadleaf L2646 ACBA 41.71 44.34166
## 3 D08 broadleaf L2645 ACBA 40.66 46.80029
## 4 D08 broadleaf P2639 ACBA 44.18 50.07397
## 5 D03 broadleaf P0614 ACFL 52.91 64.87644
## 6 D03 broadleaf L0609 ACFL 81.67 96.80924
## PLSR_CV_Predicted PLSR_CV_Residuals
## 1 53.10616 8.926162
## 2 44.39206 2.682061
## 3 46.80108 6.141077
## 4 50.16964 5.989638
## 5 65.06744 12.157436
## 6 96.86793 15.197935
cal.R2 <- round(pls::R2(plsr.out,intercept=F)[[1]][nComps],2)
cal.RMSEP <- round(sqrt(mean(cal.plsr.output$PLSR_CV_Residuals^2)),2)
val.plsr.output <- data.frame(val.plsr.data[, which(names(val.plsr.data) %notin% "Spectra")],
PLSR_Predicted=as.vector(predict(plsr.out,
newdata = val.plsr.data,
ncomp=nComps, type="response")[,,1]))
val.plsr.output <- val.plsr.output %>%
mutate(PLSR_Residuals = PLSR_Predicted-get(inVar))
head(val.plsr.output)## Domain Functional_type Sample_ID USDA_Species_Code LMA_gDW_m2 PLSR_Predicted
## 3 D02 broadleaf P0002 JUNI 60.77 64.26860
## 12 D02 broadleaf L0006 JUNI 42.54 41.53156
## 13 D02 broadleaf P0007 QUVE 106.57 99.94629
## 19 D02 broadleaf P0010 PRSE 78.82 89.09997
## 21 D02 broadleaf P0011 PRSE 86.09 84.86398
## 28 D02 broadleaf L0014 PRSE 67.11 67.88234
## PLSR_Residuals
## 3 3.4986044
## 12 -1.0084424
## 13 -6.6237126
## 19 10.2799665
## 21 -1.2260190
## 28 0.7723426
val.R2 <- round(pls::R2(plsr.out,newdata=val.plsr.data,intercept=F)[[1]][nComps],2)
val.RMSEP <- round(sqrt(mean(val.plsr.output$PLSR_Residuals^2)),2)
rng_quant <- quantile(cal.plsr.output[,inVar], probs = c(0.001, 0.999))
cal_scatter_plot <- ggplot(cal.plsr.output, aes(x=PLSR_CV_Predicted,
y=get(inVar))) +
theme_bw() + geom_point() + geom_abline(intercept = 0, slope = 1,
color="dark grey",
linetype="dashed",
linewidth=1.5) +
xlim(rng_quant[1], rng_quant[2]) +
ylim(rng_quant[1], rng_quant[2]) +
labs(x=paste0("Predicted ", paste(inVar), " (units)"),
y=paste0("Observed ", paste(inVar), " (units)"),
title=paste0("Calibration: ", paste0("Rsq = ", cal.R2), "; ",
paste0("RMSEP = ",
cal.RMSEP))) +
theme(axis.text=element_text(size=18), legend.position="none",
axis.title=element_text(size=20, face="bold"),
axis.text.x = element_text(angle = 0,vjust = 0.5),
panel.border = element_rect(linetype = "solid",
fill = NA, linewidth=1.5))
cal_resid_histogram <- ggplot(cal.plsr.output,
aes(x=PLSR_CV_Residuals)) +
geom_histogram(alpha=.5, position="identity") +
geom_vline(xintercept = 0, color="black",
linetype="dashed", linewidth=1) + theme_bw() +
theme(axis.text=element_text(size=18), legend.position="none",
axis.title=element_text(size=20, face="bold"),
axis.text.x = element_text(angle = 0,vjust = 0.5),
panel.border = element_rect(linetype = "solid",
fill = NA, linewidth=1.5))
rng_quant <- quantile(val.plsr.output[,inVar],
probs = c(0.001, 0.999))
val_scatter_plot <- ggplot(val.plsr.output,
aes(x=PLSR_Predicted, y=get(inVar))) +
theme_bw() + geom_point() +
geom_abline(intercept = 0, slope = 1, color="dark grey",
linetype="dashed", linewidth=1.5) +
xlim(rng_quant[1], rng_quant[2]) +
ylim(rng_quant[1], rng_quant[2]) +
labs(x=paste0("Predicted ", paste(inVar), " (units)"),
y=paste0("Observed ", paste(inVar), " (units)"),
title=paste0("Validation: ", paste0("Rsq = ", val.R2), "; ",
paste0("RMSEP = ",
val.RMSEP))) +
theme(axis.text=element_text(size=18), legend.position="none",
axis.title=element_text(size=20, face="bold"),
axis.text.x = element_text(angle = 0,vjust = 0.5),
panel.border = element_rect(linetype = "solid", fill = NA,
linewidth=1.5))
val_resid_histogram <- ggplot(val.plsr.output, aes(x=PLSR_Residuals)) +
geom_histogram(alpha=.5, position="identity") +
geom_vline(xintercept = 0, color="black",
linetype="dashed", linewidth=1) + theme_bw() +
theme(axis.text=element_text(size=18), legend.position="none",
axis.title=element_text(size=20, face="bold"),
axis.text.x = element_text(angle = 0,vjust = 0.5),
panel.border = element_rect(linetype = "solid", fill = NA,
linewidth=1.5))
# plot cal/val side-by-side
scatterplots <- grid.arrange(cal_scatter_plot, val_scatter_plot, cal_resid_histogram,
val_resid_histogram, nrow=2, ncol=2)## Warning: Removed 22 rows containing missing values or values outside the scale range
## (`geom_point()`).
## Warning: Removed 8 rows containing missing values or values outside the scale range
## (`geom_point()`).
## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.

ggsave(filename = file.path(outdir,paste0(inVar,"_Cal_Val_scatterplots.png")),
plot = scatterplots, device="png", width = 32, height = 30, units = "cm",
dpi = 300)vips <- spectratrait::VIP(plsr.out)[nComps,]
par(mfrow=c(2,1))
plot(plsr.out, plottype = "coef",xlab="Wavelength (nm)",
ylab="Regression coefficients",legendpos = "bottomright",
ncomp=nComps,lwd=2)
box(lwd=2.2)
plot(seq(Start.wave,End.wave,1),vips,xlab="Wavelength (nm)",ylab="VIP",cex=0.01)
lines(seq(Start.wave,End.wave,1),vips,lwd=3)
abline(h=0.8,lty=2,col="dark grey")
box(lwd=2.2)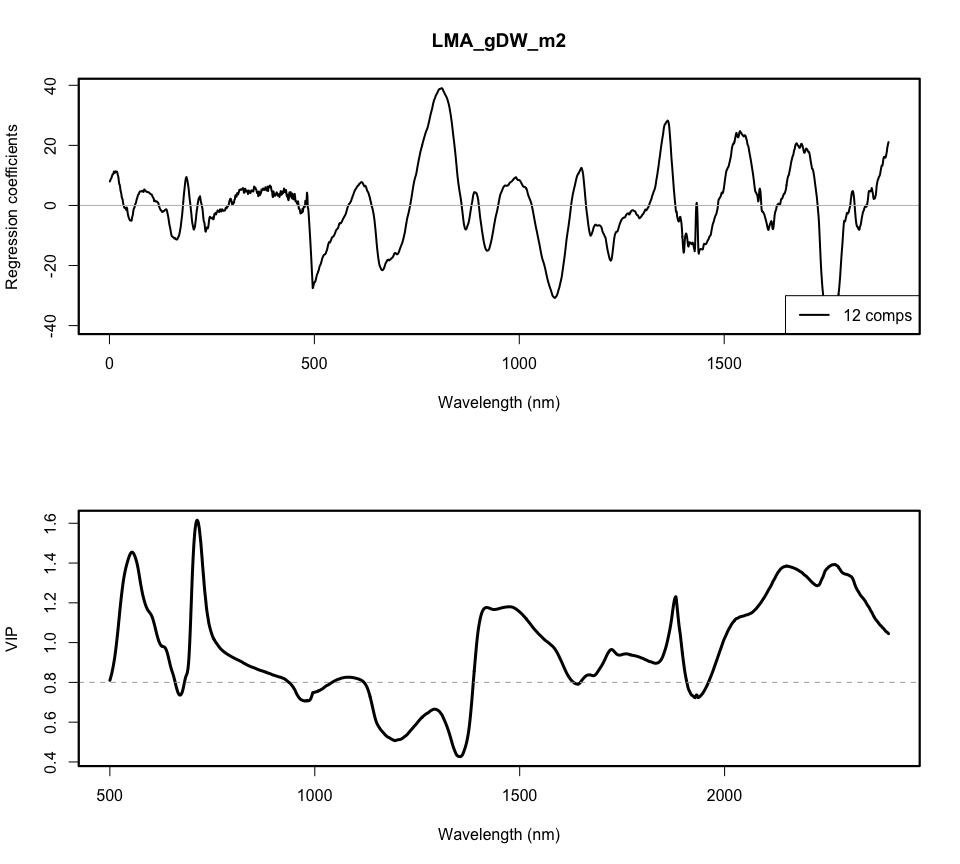
dev.copy(png,file.path(outdir,paste0(inVar,'_Coefficient_VIP_plot.png')),
height=3100, width=4100, res=340)## quartz_off_screen
## 3
dev.off();## quartz_off_screen
## 2
par(opar)if(grepl("Windows", sessionInfo()$running)){
pls.options(parallel =NULL)
} else {
pls.options(parallel = parallel::detectCores()-1)
}
seg <- 100
jk.plsr.out <- pls::plsr(as.formula(paste(inVar,"~","Spectra")), scale=FALSE,
center=TRUE, ncomp=nComps,
validation="CV", segments = seg,
segment.type="interleaved", trace=FALSE,
jackknife=TRUE, data=cal.plsr.data)
pls.options(parallel = NULL)
Jackknife_coef <- spectratrait::f.coef.valid(plsr.out = jk.plsr.out, data_plsr = cal.plsr.data,
ncomp = nComps, inVar=inVar)
Jackknife_intercept <- Jackknife_coef[1,,,]
Jackknife_coef <- Jackknife_coef[2:dim(Jackknife_coef)[1],,,]
interval <- c(0.025,0.975)
Jackknife_Pred <- val.plsr.data$Spectra %*% Jackknife_coef +
matrix(rep(Jackknife_intercept, length(val.plsr.data[,inVar])), byrow=TRUE,
ncol=length(Jackknife_intercept))
Interval_Conf <- apply(X = Jackknife_Pred,MARGIN = 1,
FUN = quantile,probs=c(interval[1],interval[2]))
sd_mean <- apply(X = Jackknife_Pred,MARGIN = 1,FUN =sd)
sd_res <- sd(val.plsr.output$PLSR_Residuals)
sd_tot <- sqrt(sd_mean^2+sd_res^2)
val.plsr.output$LCI <- Interval_Conf[1,]
val.plsr.output$UCI <- Interval_Conf[2,]
val.plsr.output$LPI <- val.plsr.output$PLSR_Predicted-1.96*sd_tot
val.plsr.output$UPI <- val.plsr.output$PLSR_Predicted+1.96*sd_tot
head(val.plsr.output)## Domain Functional_type Sample_ID USDA_Species_Code LMA_gDW_m2 PLSR_Predicted
## 3 D02 broadleaf P0002 JUNI 60.77 64.26860
## 12 D02 broadleaf L0006 JUNI 42.54 41.53156
## 13 D02 broadleaf P0007 QUVE 106.57 99.94629
## 19 D02 broadleaf P0010 PRSE 78.82 89.09997
## 21 D02 broadleaf P0011 PRSE 86.09 84.86398
## 28 D02 broadleaf L0014 PRSE 67.11 67.88234
## PLSR_Residuals LCI UCI LPI UPI
## 3 3.4986044 64.14006 64.47482 45.75155 82.78566
## 12 -1.0084424 41.40129 41.67150 23.01456 60.04855
## 13 -6.6237126 99.82564 100.08886 81.42930 118.46328
## 19 10.2799665 88.89305 89.27985 70.58206 107.61787
## 21 -1.2260190 84.71755 85.02180 66.34672 103.38124
## 28 0.7723426 67.76659 68.07148 49.36518 86.39950
spectratrait::f.plot.coef(Z = t(Jackknife_coef), wv = wv,
plot_label="Jackknife regression coefficients",position = 'bottomleft')
abline(h=0,lty=2,col="grey50")
box(lwd=2.2)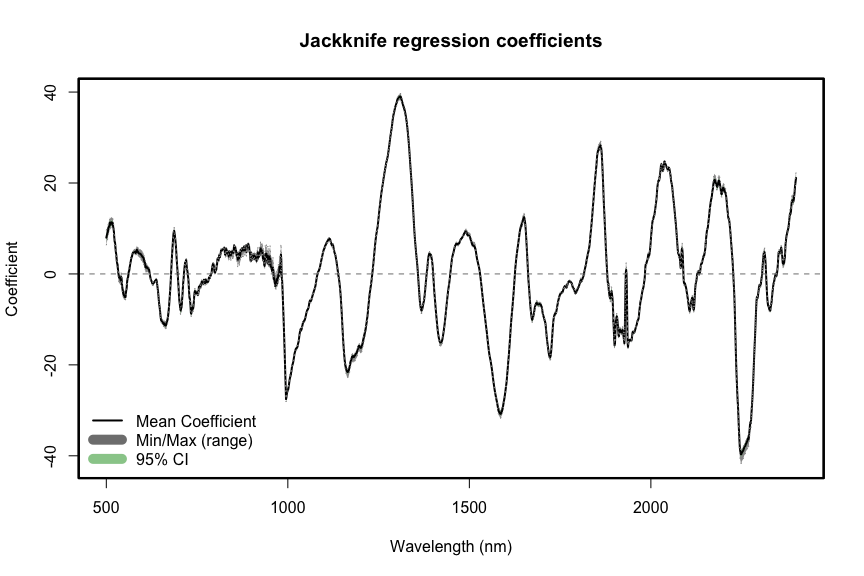
dev.copy(png,file.path(outdir,paste0(inVar,'_Jackknife_Regression_Coefficients.png')),
height=2100, width=3800, res=340)## quartz_off_screen
## 3
dev.off();## quartz_off_screen
## 2
rmsep_percrmsep <- spectratrait::percent_rmse(plsr_dataset = val.plsr.output,
inVar = inVar,
residuals = val.plsr.output$PLSR_Residuals,
range="full")
RMSEP <- rmsep_percrmsep$rmse
perc_RMSEP <- rmsep_percrmsep$perc_rmse
r2 <- round(pls::R2(plsr.out, newdata = val.plsr.data, intercept=F)$val[nComps],2)
expr <- vector("expression", 3)
expr[[1]] <- bquote(R^2==.(r2))
expr[[2]] <- bquote(RMSEP==.(round(RMSEP,2)))
expr[[3]] <- bquote("%RMSEP"==.(round(perc_RMSEP,2)))
rng_vals <- c(min(val.plsr.output$LPI), max(val.plsr.output$UPI))
par(mfrow=c(1,1), mar=c(4.2,5.3,1,0.4), oma=c(0, 0.1, 0, 0.2))
plotrix::plotCI(val.plsr.output$PLSR_Predicted,val.plsr.output[,inVar],
li=val.plsr.output$LPI, ui=val.plsr.output$UPI, gap=0.009,sfrac=0.004,
lwd=1.6, xlim=c(rng_vals[1], rng_vals[2]), ylim=c(rng_vals[1], rng_vals[2]),
err="x", pch=21, col="black", pt.bg=scales::alpha("grey70",0.7), scol="grey50",
cex=2, xlab=paste0("Predicted ", paste(inVar), " (units)"),
ylab=paste0("Observed ", paste(inVar), " (units)"),
cex.axis=1.5,cex.lab=1.8)
abline(0,1,lty=2,lw=2)
legend("topleft", legend=expr, bty="n", cex=1.5)
box(lwd=2.2)
dev.copy(png,file.path(outdir,paste0(inVar,"_PLSR_Validation_Scatterplot.png")),
height=2800, width=3200, res=340)## quartz_off_screen
## 3
dev.off();## quartz_off_screen
## 2
out.jk.coefs <- data.frame(Iteration=seq(1,seg,1),
Intercept=Jackknife_intercept,
t(Jackknife_coef))
head(out.jk.coefs)[1:6]## Iteration Intercept Wave_500 Wave_501 Wave_502 Wave_503
## Seg 1 1 68.80319 7.963544 8.372048 8.732315 8.996706
## Seg 2 2 68.33966 9.007372 9.404730 9.711349 9.942219
## Seg 3 3 67.96783 7.809066 8.218030 8.589660 8.855073
## Seg 4 4 68.56866 7.924350 8.269938 8.602341 8.843661
## Seg 5 5 68.06364 7.682055 8.080012 8.388390 8.653853
## Seg 6 6 67.73588 7.989043 8.385246 8.743061 8.962190
write.csv(out.jk.coefs,file=file.path(outdir,
paste0(inVar,
'_Jackkife_PLSR_Coefficients.csv')),
row.names=FALSE)print(paste("Output directory: ", getwd()))## [1] "Output directory: /Users/sserbin/Library/CloudStorage/OneDrive-NASA/Data/Github/spectratrait/vignettes"
# Observed versus predicted
write.csv(cal.plsr.output,file=file.path(outdir,
paste0(inVar,'_Observed_PLSR_CV_Pred_',
nComps,'comp.csv')),
row.names=FALSE)
# Validation data
write.csv(val.plsr.output,file=file.path(outdir,
paste0(inVar,'_Validation_PLSR_Pred_',
nComps,'comp.csv')),
row.names=FALSE)
# Model coefficients
coefs <- coef(plsr.out,ncomp=nComps,intercept=TRUE)
write.csv(coefs,file=file.path(outdir,
paste0(inVar,'_PLSR_Coefficients_',
nComps,'comp.csv')),
row.names=TRUE)
# PLSR VIP
write.csv(vips,file=file.path(outdir,
paste0(inVar,'_PLSR_VIPs_',
nComps,'comp.csv')))print("**** PLSR output files: ")## [1] "**** PLSR output files: "
print(list.files(outdir)[grep(pattern = inVar, list.files(outdir))])## [1] "LMA_gDW_m2_Cal_PLSR_Dataset.csv"
## [2] "LMA_gDW_m2_Cal_Val_Histograms.png"
## [3] "LMA_gDW_m2_Cal_Val_scatterplots.png"
## [4] "LMA_gDW_m2_Cal_Val_Spectra.png"
## [5] "LMA_gDW_m2_Coefficient_VIP_plot.png"
## [6] "LMA_gDW_m2_Jackkife_PLSR_Coefficients.csv"
## [7] "LMA_gDW_m2_Jackknife_Regression_Coefficients.png"
## [8] "LMA_gDW_m2_Observed_PLSR_CV_Pred_12comp.csv"
## [9] "LMA_gDW_m2_PLSR_Coefficients_12comp.csv"
## [10] "LMA_gDW_m2_PLSR_Component_Selection.png"
## [11] "LMA_gDW_m2_PLSR_Validation_Scatterplot.png"
## [12] "LMA_gDW_m2_PLSR_VIPs_12comp.csv"
## [13] "LMA_gDW_m2_Val_PLSR_Dataset.csv"
## [14] "LMA_gDW_m2_Validation_PLSR_Pred_12comp.csv"