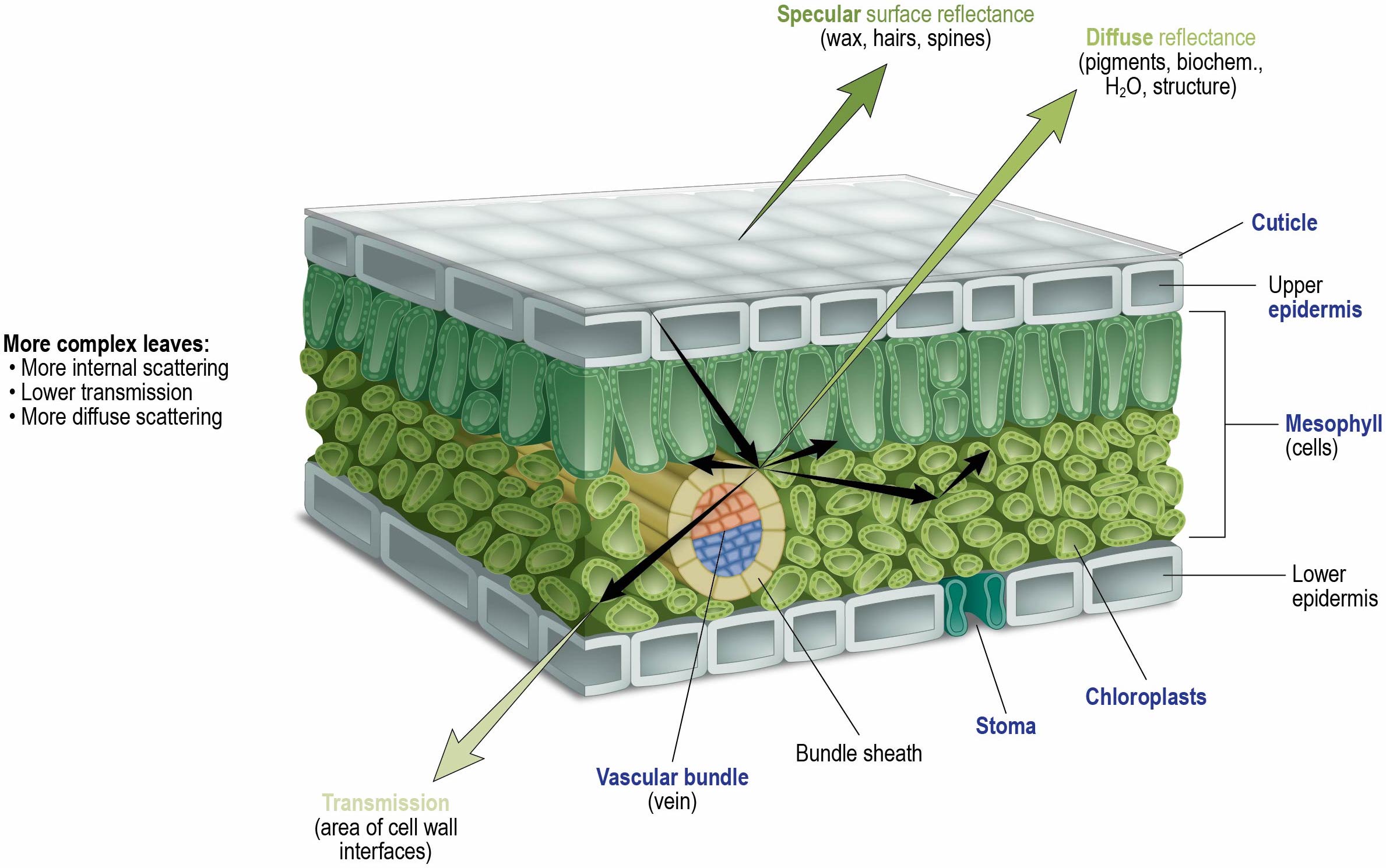
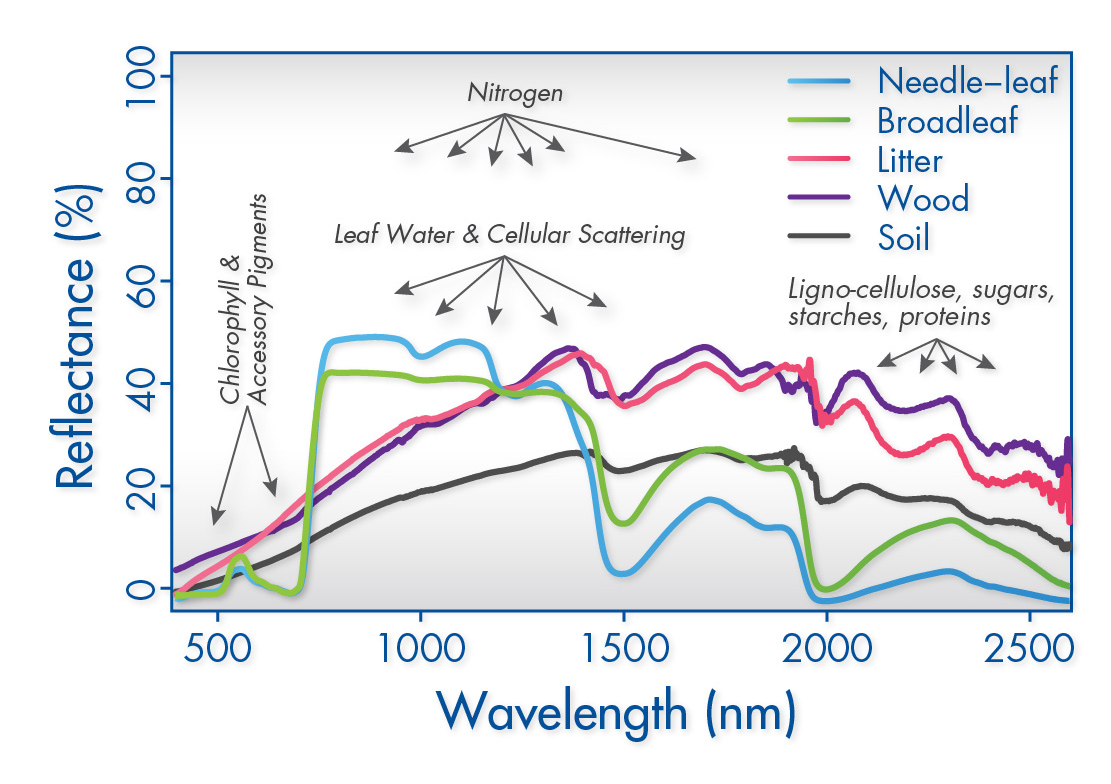
-
Notifications
You must be signed in to change notification settings - Fork 9
Example Tutorial #10: NEON CONUS dataset ‐ canopy spectra
Shawn P. Serbin edited this page Jun 19, 2024
·
1 revision
Spectra-trait PLSR example using NEON AOP pixel spectra and field-sampled leaf nitrogen content from CONUS NEON sites
Shawn P. Serbin, Julien Lamour, & Jeremiah Anderson 2024-06-19
This is an R Markdown Notebook to illustrate how to develop pixel-scale spectra-trait PLSR models. This example uses image data from NEON AOP and associated field measurements of leaf nitrogen content collected across a range of CONUS NEON sites. For more information refer to the dataset EcoSIS page: https://ecosis.org/package/canopy-spectra-to-map-foliar-functional-traits-over-neon-domains-in-eastern-united-states
list.of.packages <- c("pls","dplyr","here","plotrix","ggplot2","gridExtra","spectratrait")
invisible(lapply(list.of.packages, library, character.only = TRUE))## Warning: package 'pls' was built under R version 4.3.1
##
## Attaching package: 'pls'
## The following object is masked from 'package:stats':
##
## loadings
## Warning: package 'dplyr' was built under R version 4.3.1
##
## Attaching package: 'dplyr'
## The following objects are masked from 'package:stats':
##
## filter, lag
## The following objects are masked from 'package:base':
##
## intersect, setdiff, setequal, union
## here() starts at /Users/sserbin/Library/CloudStorage/OneDrive-NASA/Data/Github/spectratrait
## Warning: package 'plotrix' was built under R version 4.3.1
## Warning: package 'ggplot2' was built under R version 4.3.1
##
## Attaching package: 'gridExtra'
## The following object is masked from 'package:dplyr':
##
## combine
### Setup options
# Script options
pls::pls.options(plsralg = "oscorespls")
pls::pls.options("plsralg")## $plsralg
## [1] "oscorespls"
# Default par options
opar <- par(no.readonly = T)
# What is the target variable? What is the variable name in the input dataset?
inVar <- "LMA"
# What is the source dataset from EcoSIS?
ecosis_id <- "b9dbf3db-5b9c-4ab2-88c2-26c8b39d0903"
# Specify output directory, output_dir
# Options:
# tempdir - use a OS-specified temporary directory
# user defined PATH - e.g. "~/scratch/PLSR"
output_dir <- "tempdir"## [1] "/private/var/folders/th/fpt_z3417gn8xgply92pvy6r0000gq/T/Rtmprf8pzl"
print(paste0("Output directory: ",getwd())) # check wd## [1] "Output directory: /Users/sserbin/Library/CloudStorage/OneDrive-NASA/Data/Github/spectratrait/vignettes"
dat_raw <- spectratrait::get_ecosis_data(ecosis_id = ecosis_id)## [1] "**** Downloading Ecosis data ****"
## Downloading data...
## Rows: 674 Columns: 459
## ── Column specification ────────────────────────────────────────────────────────
## Delimiter: ","
## chr (4): Affiliation, PI, Plot_ID, Project
## dbl (455): Boron, Calcium, Carbon, Carotenoids_area, Carotenoids_mass, Cellu...
##
## ℹ Use `spec()` to retrieve the full column specification for this data.
## ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
## Download complete!
head(dat_raw)## # A tibble: 6 × 459
## Affiliation Boron Calcium Carbon Carotenoids_area Carotenoids_mass Cellulose
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 University … 0.0420 24.2 463. 9.19 1.18 221.
## 2 University … 0.0361 6.90 558. 10.8 1.17 183.
## 3 University … 0.0407 16.7 532. 12.2 1.52 133.
## 4 University … 0.0461 13.9 461. 9.16 1.50 220.
## 5 University … 0.0401 13.7 510. 11.0 1.53 101.
## 6 University … 0.0456 14.5 557. 8.90 1.24 214.
## # ℹ 452 more variables: Chlorophylls_area <dbl>, Chlorophylls_mass <dbl>,
## # Copper <dbl>, EWT <dbl>, Fiber <dbl>, Flavonoids <dbl>, LMA <dbl>,
## # Lignin <dbl>, Magnesium <dbl>, Manganese <dbl>, NSC <dbl>, Nitrogen <dbl>,
## # PI <chr>, Phenolics <dbl>, Phosphorus <dbl>, Plot_ID <chr>,
## # Potassium <dbl>, Project <chr>, SLA <dbl>, Sample_Year <dbl>, Starch <dbl>,
## # Sugar <dbl>, Sulfur <dbl>, Water <dbl>, d13C <dbl>, d15N <dbl>,
## # `384` <dbl>, `389` <dbl>, `394` <dbl>, `399` <dbl>, `404` <dbl>, …
names(dat_raw)[1:40]## [1] "Affiliation" "Boron" "Calcium"
## [4] "Carbon" "Carotenoids_area" "Carotenoids_mass"
## [7] "Cellulose" "Chlorophylls_area" "Chlorophylls_mass"
## [10] "Copper" "EWT" "Fiber"
## [13] "Flavonoids" "LMA" "Lignin"
## [16] "Magnesium" "Manganese" "NSC"
## [19] "Nitrogen" "PI" "Phenolics"
## [22] "Phosphorus" "Plot_ID" "Potassium"
## [25] "Project" "SLA" "Sample_Year"
## [28] "Starch" "Sugar" "Sulfur"
## [31] "Water" "d13C" "d15N"
## [34] "384" "389" "394"
## [37] "399" "404" "409"
## [40] "414"
# identify the trait data and other metadata
sample_info <- dat_raw[,names(dat_raw) %notin% seq(300,2600,1)]
head(sample_info)## # A tibble: 6 × 33
## Affiliation Boron Calcium Carbon Carotenoids_area Carotenoids_mass Cellulose
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 University … 0.0420 24.2 463. 9.19 1.18 221.
## 2 University … 0.0361 6.90 558. 10.8 1.17 183.
## 3 University … 0.0407 16.7 532. 12.2 1.52 133.
## 4 University … 0.0461 13.9 461. 9.16 1.50 220.
## 5 University … 0.0401 13.7 510. 11.0 1.53 101.
## 6 University … 0.0456 14.5 557. 8.90 1.24 214.
## # ℹ 26 more variables: Chlorophylls_area <dbl>, Chlorophylls_mass <dbl>,
## # Copper <dbl>, EWT <dbl>, Fiber <dbl>, Flavonoids <dbl>, LMA <dbl>,
## # Lignin <dbl>, Magnesium <dbl>, Manganese <dbl>, NSC <dbl>, Nitrogen <dbl>,
## # PI <chr>, Phenolics <dbl>, Phosphorus <dbl>, Plot_ID <chr>,
## # Potassium <dbl>, Project <chr>, SLA <dbl>, Sample_Year <dbl>, Starch <dbl>,
## # Sugar <dbl>, Sulfur <dbl>, Water <dbl>, d13C <dbl>, d15N <dbl>
# spectra matrix
Spectra <- as.matrix(dat_raw[,names(dat_raw) %notin% names(sample_info)])
# set the desired spectra wavelength range to include
Start.wave <- 500
End.wave <- 2400
wv <- seq(Start.wave,End.wave,1)
final_spec <- Spectra[,round(as.numeric(colnames(Spectra))) %in% wv]
colnames(final_spec) <- c(paste0("Wave_",colnames(final_spec)))
## Drop bad spectra data - for canopy-scale reflectance, often the "water band" wavelengths
## are too noisy to use for trait estimation. Its possible to remove these wavelengths
## prior to model fitting. Its best to first identify which wavelengths to drop
## before attempting PLSR, as these ranges may need to be considered on a case-by-case
## basis or generalized for multiple datasets
dropwaves <- c(1350:1440, 1826:1946)
final_spec <- final_spec[,colnames(final_spec) %notin% paste0("Wave_",dropwaves)]
wv <- as.numeric(gsub(pattern = "Wave_",replacement = "", x = colnames(final_spec)))
## Drop bad spectra data - for canopy-scale reflectance, often the "water band" wavelengths
## are too noisy to use for trait estimation. Its possible to remove these wavelengths
## prior to model fitting. Its best to first identify which wavelengths to drop
## before attempting PLSR, as these ranges may need to be considered on a case-by-case
## basis or generalized for multiple datasets
dropwaves <- c(1350:1440, 1826:1946)
final_spec <- final_spec[,colnames(final_spec) %notin% paste0("Wave_",dropwaves)]
wv <- as.numeric(gsub(pattern = "Wave_",replacement = "", x = colnames(final_spec)))
# assemble example dataset - !!can add more traits here to try developing additional models
sample_info2 <- sample_info %>%
select(Plot_ID,Sample_Year,SLA,LMA,Nitrogen)
site_plot <- data.frame(matrix(unlist(strsplit(sample_info2$Plot_ID, "_")),
ncol=2, byrow=TRUE))
colnames(site_plot) <- c("Plot_Num","SampleID")
sample_info3 <- data.frame(site_plot,sample_info2)
plsr_data <- data.frame(sample_info3,final_spec*0.01)
rm(sample_info,sample_info2,sample_info3,Spectra, site_plot)# Example data cleaning. End user needs to do what's appropriate for their
# data. This may be an iterative process.
# Keep only complete rows of inVar and spec data before fitting
#
plsr_data <- plsr_data %>% # remove erroneously high values, or "bad spectra"
filter(Nitrogen<50) %>%
filter(Wave_859<80) %>%
filter(Wave_859>15)
plsr_data <- plsr_data[complete.cases(plsr_data[,names(plsr_data) %in%
c(inVar,paste0("Wave_",wv))]),]## Make a stratified random sampling in the strata USDA_Species_Code and Domain
method <- "base" #base/dplyr
# base R - a bit slow
# dplyr - much faster
split_data <- spectratrait::create_data_split(dataset=plsr_data, approach=method, split_seed=2356326,
prop=0.8, group_variables="Plot_Num")## D02 Cal: 80.46%
## D03 Cal: 80.328%
## D05 Cal: 80%
## D06 Cal: 80.137%
## D07 Cal: 79.245%
## D08 Cal: 79.817%
## D09 Cal: 79.63%
names(split_data)## [1] "cal_data" "val_data"
cal.plsr.data <- split_data$cal_data
head(cal.plsr.data)[1:8]## Plot_Num SampleID Plot_ID Sample_Year SLA LMA Nitrogen Wave_504
## 2 D02 0002 D02_0002 2017 10.77861 92.77636 27.70598 1.2909576
## 3 D02 0003 D02_0003 2017 12.46154 80.24688 34.63999 1.2976806
## 5 D02 0005 D02_0005 2017 17.27620 57.88311 26.64623 1.7735714
## 6 D02 0006 D02_0006 2017 12.92806 77.35113 20.69437 1.7786337
## 7 D02 0007 D02_0007 2017 10.21521 97.89327 28.87526 1.7981043
## 8 D02 0008 D02_0008 2017 20.87397 47.90656 33.63137 0.8780127
val.plsr.data <- split_data$val_data
head(val.plsr.data)[1:8]## Plot_Num SampleID Plot_ID Sample_Year SLA LMA Nitrogen Wave_504
## 1 D02 0001 D02_0001 2017 13.66366 73.18685 31.18030 1.467240
## 4 D02 0004 D02_0004 2017 16.63205 60.12487 34.54034 1.551933
## 16 D02 0016 D02_0016 2017 14.44765 69.21540 22.87740 2.198174
## 18 D02 0019 D02_0019 2017 14.47103 69.10360 17.73126 1.961911
## 19 D02 0020 D02_0020 2017 18.98522 52.67256 21.32929 1.546430
## 20 D02 0021 D02_0021 2017 12.12731 82.45849 29.50256 1.936263
rm(split_data)
# Datasets:
print(paste("Cal observations: ",dim(cal.plsr.data)[1],sep=""))## [1] "Cal observations: 516"
print(paste("Val observations: ",dim(val.plsr.data)[1],sep=""))## [1] "Val observations: 129"
cal_hist_plot <- ggplot(data = cal.plsr.data,
aes(x = cal.plsr.data[,paste0(inVar)])) +
geom_histogram(fill=I("grey50"),col=I("black"),alpha=I(.7)) +
labs(title=paste0("Calibration Histogram for ",inVar), x = paste0(inVar),
y = "Count")
val_hist_plot <- ggplot(data = val.plsr.data,
aes(x = val.plsr.data[,paste0(inVar)])) +
geom_histogram(fill=I("grey50"),col=I("black"),alpha=I(.7)) +
labs(title=paste0("Validation Histogram for ",inVar), x = paste0(inVar),
y = "Count")
histograms <- grid.arrange(cal_hist_plot, val_hist_plot, ncol=2)## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.

ggsave(filename = file.path(outdir,paste0(inVar,"_Cal_Val_Histograms.png")), plot = histograms,
device="png", width = 30,
height = 12, units = "cm",
dpi = 300)
# output cal/val data
write.csv(cal.plsr.data,file=file.path(outdir,paste0(inVar,'_Cal_PLSR_Dataset.csv')),
row.names=FALSE)
write.csv(val.plsr.data,file=file.path(outdir,paste0(inVar,'_Val_PLSR_Dataset.csv')),
row.names=FALSE)cal_spec <- as.matrix(cal.plsr.data[, which(names(cal.plsr.data) %in% paste0("Wave_",wv))])
cal.plsr.data <- data.frame(cal.plsr.data[, which(names(cal.plsr.data) %notin% paste0("Wave_",wv))],
Spectra=I(cal_spec))
head(cal.plsr.data)[1:5]## Plot_Num SampleID Plot_ID Sample_Year SLA
## 2 D02 0002 D02_0002 2017 10.77861
## 3 D02 0003 D02_0003 2017 12.46154
## 5 D02 0005 D02_0005 2017 17.27620
## 6 D02 0006 D02_0006 2017 12.92806
## 7 D02 0007 D02_0007 2017 10.21521
## 8 D02 0008 D02_0008 2017 20.87397
val_spec <- as.matrix(val.plsr.data[, which(names(val.plsr.data) %in% paste0("Wave_",wv))])
val.plsr.data <- data.frame(val.plsr.data[, which(names(val.plsr.data) %notin% paste0("Wave_",wv))],
Spectra=I(val_spec))
head(val.plsr.data)[1:5]## Plot_Num SampleID Plot_ID Sample_Year SLA
## 1 D02 0001 D02_0001 2017 13.66366
## 4 D02 0004 D02_0004 2017 16.63205
## 16 D02 0016 D02_0016 2017 14.44765
## 18 D02 0019 D02_0019 2017 14.47103
## 19 D02 0020 D02_0020 2017 18.98522
## 20 D02 0021 D02_0021 2017 12.12731
par(mfrow=c(1,2)) # B, L, T, R
spectratrait::f.plot.spec(Z=cal.plsr.data$Spectra,wv=wv,plot_label="Calibration")
spectratrait::f.plot.spec(Z=val.plsr.data$Spectra,wv=wv,plot_label="Validation")
dev.copy(png,file.path(outdir,paste0(inVar,'_Cal_Val_Spectra.png')),
height=2500,width=4900, res=340)## quartz_off_screen
## 3
dev.off();## quartz_off_screen
## 2
par(mfrow=c(1,1))if(grepl("Windows", sessionInfo()$running)){
pls.options(parallel = NULL)
} else {
pls.options(parallel = parallel::detectCores()-1)
}
method <- "pls" #pls, firstPlateau, firstMin
random_seed <- 1245565
seg <- 50
maxComps <- 16
iterations <- 80
prop <- 0.70
if (method=="pls") {
# pls package approach - faster but estimates more components....
nComps <- spectratrait::find_optimal_components(dataset=cal.plsr.data, targetVariable=inVar,
method=method,
maxComps=maxComps, seg=seg,
random_seed=random_seed)
print(paste0("*** Optimal number of components: ", nComps))
} else {
nComps <- spectratrait::find_optimal_components(dataset=cal.plsr.data, targetVariable=inVar,
method=method,
maxComps=maxComps, iterations=iterations,
seg=seg, prop=prop,
random_seed=random_seed)
}## [1] "*** Identifying optimal number of PLSR components ***"
## [1] "*** Running PLS permutation test ***"

## [1] "*** Optimal number of components: 13"
dev.copy(png,file.path(outdir,paste0(paste0(inVar,"_PLSR_Component_Selection.png"))),
height=2800, width=3400, res=340)## quartz_off_screen
## 3
dev.off();## quartz_off_screen
## 2
plsr.out <- plsr(as.formula(paste(inVar,"~","Spectra")),scale=FALSE,ncomp=nComps,validation="LOO",
trace=FALSE,data=cal.plsr.data)
fit <- plsr.out$fitted.values[,1,nComps]
pls.options(parallel = NULL)
# External validation fit stats
par(mfrow=c(1,2)) # B, L, T, R
pls::RMSEP(plsr.out, newdata = val.plsr.data)## (Intercept) 1 comps 2 comps 3 comps 4 comps 5 comps
## 63.00 53.46 52.75 48.09 45.07 44.69
## 6 comps 7 comps 8 comps 9 comps 10 comps 11 comps
## 40.32 40.35 39.18 38.26 36.82 35.26
## 12 comps 13 comps
## 35.26 33.94
plot(pls::RMSEP(plsr.out,estimate=c("test"),newdata = val.plsr.data), main="MODEL RMSEP",
xlab="Number of Components",ylab="Model Validation RMSEP",lty=1,col="black",cex=1.5,lwd=2)
box(lwd=2.2)
R2(plsr.out, newdata = val.plsr.data)## (Intercept) 1 comps 2 comps 3 comps 4 comps 5 comps
## -4.712e-05 2.798e-01 2.987e-01 4.173e-01 4.882e-01 4.968e-01
## 6 comps 7 comps 8 comps 9 comps 10 comps 11 comps
## 5.903e-01 5.898e-01 6.132e-01 6.311e-01 6.583e-01 6.867e-01
## 12 comps 13 comps
## 6.867e-01 7.098e-01
plot(pls::R2(plsr.out,estimate=c("test"),newdata = val.plsr.data), main="MODEL R2",
xlab="Number of Components",ylab="Model Validation R2",lty=1,col="black",cex=1.5,lwd=2)
box(lwd=2.2)
dev.copy(png,file.path(outdir,paste0(paste0(inVar,"_Validation_RMSEP_R2_by_Component.png"))),
height=2800, width=4800, res=340)## quartz_off_screen
## 3
dev.off();## quartz_off_screen
## 2
par(opar)#calibration
cal.plsr.output <- data.frame(cal.plsr.data[, which(names(cal.plsr.data) %notin% "Spectra")],
PLSR_Predicted=fit,
PLSR_CV_Predicted=as.vector(plsr.out$validation$pred[,,nComps]))
cal.plsr.output <- cal.plsr.output %>%
mutate(PLSR_CV_Residuals = PLSR_CV_Predicted-get(inVar))
head(cal.plsr.output)## Plot_Num SampleID Plot_ID Sample_Year SLA LMA Nitrogen
## 2 D02 0002 D02_0002 2017 10.77861 92.77636 27.70598
## 3 D02 0003 D02_0003 2017 12.46154 80.24688 34.63999
## 5 D02 0005 D02_0005 2017 17.27620 57.88311 26.64623
## 6 D02 0006 D02_0006 2017 12.92806 77.35113 20.69437
## 7 D02 0007 D02_0007 2017 10.21521 97.89327 28.87526
## 8 D02 0008 D02_0008 2017 20.87397 47.90656 33.63137
## PLSR_Predicted PLSR_CV_Predicted PLSR_CV_Residuals
## 2 103.49368 103.69951 10.923148
## 3 89.25497 89.41222 9.165342
## 5 27.39939 25.44901 -32.434102
## 6 100.42843 100.91913 23.568000
## 7 118.92179 119.82453 21.931263
## 8 71.24724 71.99274 24.086180
cal.R2 <- round(pls::R2(plsr.out,intercept=F)[[1]][nComps],2)
cal.RMSEP <- round(sqrt(mean(cal.plsr.output$PLSR_CV_Residuals^2)),2)
val.plsr.output <- data.frame(val.plsr.data[, which(names(val.plsr.data) %notin% "Spectra")],
PLSR_Predicted=as.vector(predict(plsr.out,
newdata = val.plsr.data,
ncomp=nComps, type="response")[,,1]))
val.plsr.output <- val.plsr.output %>%
mutate(PLSR_Residuals = PLSR_Predicted-get(inVar))
head(val.plsr.output)## Plot_Num SampleID Plot_ID Sample_Year SLA LMA Nitrogen
## 1 D02 0001 D02_0001 2017 13.66366 73.18685 31.18030
## 4 D02 0004 D02_0004 2017 16.63205 60.12487 34.54034
## 16 D02 0016 D02_0016 2017 14.44765 69.21540 22.87740
## 18 D02 0019 D02_0019 2017 14.47103 69.10360 17.73126
## 19 D02 0020 D02_0020 2017 18.98522 52.67256 21.32929
## 20 D02 0021 D02_0021 2017 12.12731 82.45849 29.50256
## PLSR_Predicted PLSR_Residuals
## 1 88.88277 15.695919
## 4 31.24382 -28.881047
## 16 72.11545 2.900049
## 18 94.05676 24.953162
## 19 67.49522 14.822652
## 20 84.50151 2.043023
val.R2 <- round(pls::R2(plsr.out,newdata=val.plsr.data,intercept=F)[[1]][nComps],2)
val.RMSEP <- round(sqrt(mean(val.plsr.output$PLSR_Residuals^2)),2)
rng_quant <- quantile(cal.plsr.output[,inVar], probs = c(0.001, 0.999))
cal_scatter_plot <- ggplot(cal.plsr.output, aes(x=PLSR_CV_Predicted, y=get(inVar))) +
theme_bw() + geom_point() + geom_abline(intercept = 0, slope = 1, color="dark grey",
linetype="dashed", linewidth=1.5) +
xlim(rng_quant[1], rng_quant[2]) +
ylim(rng_quant[1], rng_quant[2]) +
labs(x=paste0("Predicted ", paste(inVar), " (units)"),
y=paste0("Observed ", paste(inVar), " (units)"),
title=paste0("Calibration: ", paste0("Rsq = ", cal.R2), "; ", paste0("RMSEP = ",
cal.RMSEP))) +
theme(axis.text=element_text(size=18), legend.position="none",
axis.title=element_text(size=20, face="bold"),
axis.text.x = element_text(angle = 0,vjust = 0.5),
panel.border = element_rect(linetype = "solid", fill = NA, linewidth=1.5))
cal_resid_histogram <- ggplot(cal.plsr.output, aes(x=PLSR_CV_Residuals)) +
geom_histogram(alpha=.5, position="identity") +
geom_vline(xintercept = 0, color="black",
linetype="dashed", linewidth=1) + theme_bw() +
theme(axis.text=element_text(size=18), legend.position="none",
axis.title=element_text(size=20, face="bold"),
axis.text.x = element_text(angle = 0,vjust = 0.5),
panel.border = element_rect(linetype = "solid", fill = NA, linewidth=1.5))
rng_quant <- quantile(val.plsr.output[,inVar], probs = c(0.001, 0.999))
val_scatter_plot <- ggplot(val.plsr.output, aes(x=PLSR_Predicted, y=get(inVar))) +
theme_bw() + geom_point() + geom_abline(intercept = 0, slope = 1, color="dark grey",
linetype="dashed", linewidth=1.5) +
xlim(rng_quant[1], rng_quant[2]) +
ylim(rng_quant[1], rng_quant[2]) +
labs(x=paste0("Predicted ", paste(inVar), " (units)"),
y=paste0("Observed ", paste(inVar), " (units)"),
title=paste0("Validation: ", paste0("Rsq = ", val.R2), "; ", paste0("RMSEP = ",
val.RMSEP))) +
theme(axis.text=element_text(size=18), legend.position="none",
axis.title=element_text(size=20, face="bold"),
axis.text.x = element_text(angle = 0,vjust = 0.5),
panel.border = element_rect(linetype = "solid", fill = NA, linewidth=1.5))
val_resid_histogram <- ggplot(val.plsr.output, aes(x=PLSR_Residuals)) +
geom_histogram(alpha=.5, position="identity") +
geom_vline(xintercept = 0, color="black",
linetype="dashed", size=1) + theme_bw() +
theme(axis.text=element_text(size=18), legend.position="none",
axis.title=element_text(size=20, face="bold"),
axis.text.x = element_text(angle = 0,vjust = 0.5),
panel.border = element_rect(linetype = "solid", fill = NA, size=1.5))## Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
## ℹ Please use `linewidth` instead.
## This warning is displayed once every 8 hours.
## Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
## generated.
## Warning: The `size` argument of `element_rect()` is deprecated as of ggplot2 3.4.0.
## ℹ Please use the `linewidth` argument instead.
## This warning is displayed once every 8 hours.
## Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
## generated.
# plot cal/val side-by-side
scatterplots <- grid.arrange(cal_scatter_plot, val_scatter_plot, cal_resid_histogram,
val_resid_histogram, nrow=2,ncol=2)## Warning: Removed 22 rows containing missing values or values outside the scale range
## (`geom_point()`).
## Warning: Removed 8 rows containing missing values or values outside the scale range
## (`geom_point()`).
## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.

ggsave(filename = file.path(outdir,paste0(inVar,"_Cal_Val_Scatterplots.png")),
plot = scatterplots, device="png", width = 32, height = 30, units = "cm",
dpi = 300)vips <- spectratrait::VIP(plsr.out)[nComps,]
par(mfrow=c(2,1))
plot(plsr.out$coefficients[,,nComps], x=wv,xlab="Wavelength (nm)",
ylab="Regression coefficients",lwd=2,type='l')
box(lwd=2.2)
plot(wv, vips, xlab="Wavelength (nm)",ylab="VIP",cex=0.01)
lines(wv, vips, lwd=3)
abline(h=0.8, lty=2, col="dark grey")
box(lwd=2.2)
dev.copy(png, file.path(outdir, paste0(inVar,'_Coefficient_VIP_plot.png')),
height=3100, width=4100, res=340)## quartz_off_screen
## 3
dev.off();## quartz_off_screen
## 2
par(opar)## [1] "*** Running permutation test. Please hang tight, this can take awhile ***"
## [1] "Options:"
## [1] "Max Components: 13 Iterations: 500 Data Proportion (percent): 70"
## [1] "*** Providing PRESS and coefficient array output ***"
## Plot_Num SampleID Plot_ID Sample_Year SLA LMA Nitrogen
## 1 D02 0001 D02_0001 2017 13.66366 73.18685 31.18030
## 4 D02 0004 D02_0004 2017 16.63205 60.12487 34.54034
## 16 D02 0016 D02_0016 2017 14.44765 69.21540 22.87740
## 18 D02 0019 D02_0019 2017 14.47103 69.10360 17.73126
## 19 D02 0020 D02_0020 2017 18.98522 52.67256 21.32929
## 20 D02 0021 D02_0021 2017 12.12731 82.45849 29.50256
## PLSR_Predicted PLSR_Residuals LCI UCI LPI UPI
## 1 88.88277 15.695919 80.31958 94.78715 21.91528946 155.8502
## 4 31.24382 -28.881047 20.11453 40.56775 -36.24765589 98.7353
## 16 72.11545 2.900049 56.63684 79.25216 4.53409981 139.6968
## 18 94.05676 24.953162 83.62375 101.86102 26.84773695 161.2658
## 19 67.49522 14.822652 56.21051 78.44636 0.01292422 134.9775
## 20 84.50151 2.043023 67.56962 96.22524 16.47738989 152.5256
spectratrait::f.plot.coef(Z = t(bootstrap_coef), wv = wv,
plot_label="Bootstrap regression coefficients",
position = 'bottomleft')
abline(h=0,lty=2,col="grey50")
box(lwd=2.2)
dev.copy(png,file.path(outdir,paste0(inVar,'_Bootstrap_Regression_Coefficients.png')),
height=2100, width=3800, res=340)## quartz_off_screen
## 3
dev.off();## quartz_off_screen
## 2
rmsep_percrmsep <- spectratrait::percent_rmse(plsr_dataset = val.plsr.output,
inVar = inVar,
residuals = val.plsr.output$PLSR_Residuals,
range="full")
RMSEP <- rmsep_percrmsep$rmse
perc_RMSEP <- rmsep_percrmsep$perc_rmse
r2 <- round(pls::R2(plsr.out, newdata = val.plsr.data, intercept=F)$val[nComps],2)
expr <- vector("expression", 3)
expr[[1]] <- bquote(R^2==.(r2))
expr[[2]] <- bquote(RMSEP==.(round(RMSEP,2)))
expr[[3]] <- bquote("%RMSEP"==.(round(perc_RMSEP,2)))
rng_vals <- c(min(val.plsr.output$LPI), max(val.plsr.output$UPI))
par(mfrow=c(1,1), mar=c(4.2,5.3,1,0.4), oma=c(0, 0.1, 0, 0.2))
plotrix::plotCI(val.plsr.output$PLSR_Predicted,val.plsr.output[,inVar],
li=val.plsr.output$LPI, ui=val.plsr.output$UPI, gap=0.009,sfrac=0.000,
lwd=1.6, xlim=c(rng_vals[1], rng_vals[2]), ylim=c(rng_vals[1], rng_vals[2]),
err="x", pch=21, col="black", pt.bg=scales::alpha("grey70",0.7), scol="grey80",
cex=2, xlab=paste0("Predicted ", paste(inVar), " (units)"),
ylab=paste0("Observed ", paste(inVar), " (units)"),
cex.axis=1.5,cex.lab=1.8)
abline(0,1,lty=2,lw=2)
plotrix::plotCI(val.plsr.output$PLSR_Predicted,val.plsr.output[,inVar],
li=val.plsr.output$LCI, ui=val.plsr.output$UCI, gap=0.009,sfrac=0.004,
lwd=1.6, xlim=c(rng_vals[1], rng_vals[2]), ylim=c(rng_vals[1], rng_vals[2]),
err="x", pch=21, col="black", pt.bg=scales::alpha("grey70",0.7), scol="black",
cex=2, xlab=paste0("Predicted ", paste(inVar), " (units)"),
ylab=paste0("Observed ", paste(inVar), " (units)"),
cex.axis=1.5,cex.lab=1.8, add=T)
legend("topleft", legend=expr, bty="n", cex=1.5)
legend("bottomright", legend=c("Prediction Interval","Confidence Interval"),
lty=c(1,1), col = c("grey80","black"), lwd=3, bty="n", cex=1.5)
box(lwd=2.2)
dev.copy(png,file.path(outdir,paste0(inVar,"_PLSR_Validation_Scatterplot.png")),
height=2800, width=3200, res=340)## quartz_off_screen
## 3
dev.off();## quartz_off_screen
## 2
out.jk.coefs <- data.frame(Iteration=seq(1,length(bootstrap_intercept),1),
Intercept=bootstrap_intercept,t(bootstrap_coef))
names(out.jk.coefs) <- c("Iteration","Intercept",paste0("Wave_",wv))
head(out.jk.coefs)[1:6]## Iteration Intercept Wave_504 Wave_509 Wave_514 Wave_519
## 1 1 142.6449 5.389061 5.063662 4.549220 3.934070
## 2 2 136.3290 5.778862 5.071949 4.480130 3.802164
## 3 3 137.9760 6.557700 5.852310 5.098870 4.180933
## 4 4 134.0813 4.481368 4.064646 3.625608 3.325396
## 5 5 150.0042 4.412705 4.119657 3.758452 3.367088
## 6 6 121.3683 7.114084 6.469596 5.689691 4.846154
write.csv(out.jk.coefs,file=file.path(outdir,paste0(inVar,'_Bootstrap_PLSR_Coefficients.csv')),
row.names=FALSE)print(paste("Output directory: ", getwd()))## [1] "Output directory: /Users/sserbin/Library/CloudStorage/OneDrive-NASA/Data/Github/spectratrait/vignettes"
# Observed versus predicted
write.csv(cal.plsr.output,file=file.path(outdir,
paste0(inVar,'_Observed_PLSR_CV_Pred_',nComps,
'comp.csv')),row.names=FALSE)
# Validation data
write.csv(val.plsr.output,file=file.path(outdir,
paste0(inVar,'_Validation_PLSR_Pred_',nComps,
'comp.csv')),row.names=FALSE)
# Model coefficients
coefs <- coef(plsr.out,ncomp=nComps,intercept=TRUE)
write.csv(coefs,file=file.path(outdir,paste0(inVar,'_PLSR_Coefficients_',
nComps,'comp.csv')),
row.names=TRUE)
# PLSR VIP
write.csv(vips,file=file.path(outdir,paste0(inVar,
'_PLSR_VIPs_',nComps,
'comp.csv')))print("**** PLSR output files: ")## [1] "**** PLSR output files: "
print(list.files(outdir)[grep(pattern = inVar,
list.files(outdir))])## [1] "LMA_Bootstrap_PLSR_Coefficients.csv"
## [2] "LMA_Bootstrap_Regression_Coefficients.png"
## [3] "LMA_Cal_PLSR_Dataset.csv"
## [4] "LMA_Cal_Val_Histograms.png"
## [5] "LMA_Cal_Val_Scatterplots.png"
## [6] "LMA_Cal_Val_Spectra.png"
## [7] "LMA_Coefficient_VIP_plot.png"
## [8] "LMA_Observed_PLSR_CV_Pred_13comp.csv"
## [9] "LMA_PLSR_Coefficients_13comp.csv"
## [10] "LMA_PLSR_Component_Selection.png"
## [11] "LMA_PLSR_Validation_Scatterplot.png"
## [12] "LMA_PLSR_VIPs_13comp.csv"
## [13] "LMA_Val_PLSR_Dataset.csv"
## [14] "LMA_Validation_PLSR_Pred_13comp.csv"
## [15] "LMA_Validation_RMSEP_R2_by_Component.png"